Gut Health, Well-Being, and More: The Essential Role of Serotonin in the Body
Serotonin, popularly known as the “happiness hormone,” is a fundamental…
Continue reading

In recent times, the number of people experiencing discomfort related to the digestive system is growing. Even without severe symptoms, this often affects the quality of life. The gut microbiome, with all its components and interactions, forms a complete ecological niche that interacts with our physiology.
Today’s lifestyle, with high stress levels, inadequate diet or certain treatments, can alter the gut microbiome, causing an imbalance in the various types of microorganisms inhabiting our gut, which can negatively impact our health. Therefore, achieving a balanced microbiome is essential for good health (1).
The term microbiome refers to the collective genome of a microbial community; it is the complete set of genes of microorganisms forming the microbiota, which includes the genetic material of all gut bacteria, viruses, fungi, and other microorganisms.
Additionally, the term also refers to the metabolic activities and capabilities of the present microorganisms (2), such as the metabolites produced and consumed by these microorganisms.
The gut microbiota represents a complex ecosystem that develops alongside its hosts and depends on their physiological environment. The gut bacterial population stabilizes during the first years of life and remains stable throughout our lives, dominated by four main phyla: Firmicutes, Bacteroidetes, Actinobacteria and Proteobacteria.
The number of microbial species in the human intestine is estimated to be between 1,000 and 1,150, with the number of genes in the intestinal microbiome exceeding the number of genes in the human genome by 150 times (3).
The gut microbiota can perform many processes that the host cannot. These processes result in microbially produced or modulated metabolites that act as metabolic substrates and signaling molecules in the host, with significant implications for metabolism and health.
Dietary composition is fundamental for the metabolic production of the gut microbiota, as it processes dietary nutrients into metabolites. Thus, diet affects the composition of the gut microbiota, influencing its metabolic potential and impact on the host (4).
The microbiota acquired at birth develops in parallel with the host’s development, maintaining its temporal stability and diversity throughout adult life until death.
The literature suggests that although part of the microbiota is conserved, dynamic members vary throughout the gastrointestinal tract from infancy to old age under different health conditions.
Despite being dynamic, the gut microbiota performs several basic functions in the immunological, metabolic, structural, and neurological scenarios of the human body.
It significantly influences both the physical and mental health of individuals (5), regulating many metabolic processes in the host, including energy homeostasis, glucose metabolism, and lipid metabolismo (6).
The gut microbiota functions in four distinct contexts in the human body (7):
The complex communities of microorganisms inhabiting the human gastrointestinal tract are emerging as key elements in regulating health and disease. Various vital functions performed by the gut microbiome in the human body highlight its importance.
These include transforming indigestible food parts into absorbable substances, producing essential vitamins, eliminating toxic substances, competing with pathogens, strengthening the intestinal barrier, and regulating the immune system.
Many of these functions are closely linked to human physiology. For example, short-chain fatty acids produced by microbial fermentation are crucial for intestinal cells and play essential roles in regulating the immune system (8-10).
In the early stages of life, microbiome development influences immune function. Microorganisms acquired vertically, horizontally, and from the environment, as well as their metabolic products, have the potential to shape developmental courses impacting health throughout life (11).
Thus, the microbiome plays a crucial role in developing metabolic, immunological, and nutritional functions, necessitating careful attention. Understanding how complex microbial communities can affect the pathogenesis of various diseases has significant implications for their prevention, diagnosis, and treatment (12).
In recent decades, human microbiome research has evolved beyond merely cataloging microbial diversity to understanding how these microorganisms constitute an auxiliary and dynamic functional system that develops synergistically alongside physiological development and decline (13-15).
It is increasingly evident that a wide range of conditions, including chronic inflammatory diseases (16), metabolic diseases (17), neurological disorders and câncer (18, 19), are now linked to functional changes in the microbiome.
These changes can occur both at the disease manifestation site and in distant mucosal areas or organ systems, triggering metabolic and immunological changes in the host (20).
Various factors such as diet, antimicrobial agents, and immunity influence human microbiomes, particularly the gut microbiome, which houses the largest quantity and variety of microorganisms. In response, bioactive products from the microbiome shape human cell function (2, 21).
After reaching the age of two to three, the gut microbiome becomes more stable and, at this stage, closely resembles an adult microbiome (20). However, the adult gut microbiome can still be influenced by many factors, with diet being the most influential, followed by medications.
Stress also plays a significant role as the brain can influence the gut microbiome by sending stress hormones to the gut through the vagus nerve. Exercise habits, age, genetics, geographic location, and ethnicity are other elements that impact the gut microbiome (16).
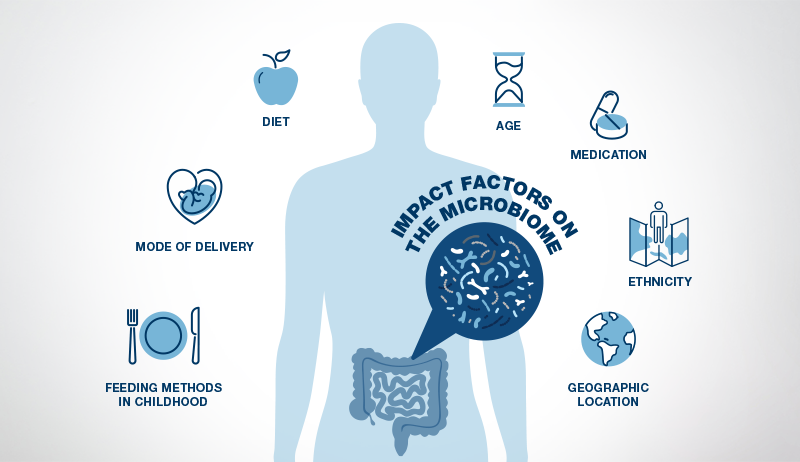
Imbalances in the gut microbiome can be qualitative or quantitative, affecting both its distribution and activity, which has a proven effect on intestinal and overall health.
Diseases related to gut microbiome imbalance include conditions such as:
The mechanism of this relationship stems from the fact that microorganisms can produce different biologically active metabolites, which can play a role in disease states.
The causality between the gut and each disease is still being established, and it is unknown whether the disease causes changes in the gut microbiome or a change in the gut microbiome causes the disease.
Several intestinal diseases linked to inflammation have been correlated with changes in the gut microbiome. These include diseases such as colon cancer, ulcerative colitis, Crohn’s disease, and irritable bowel syndrome (3, 22, 23).
Crohn’s disease (CD) is an inflammatory bowel disease (IBD) that can affect any part of the gastrointestinal tract. CD has been related to functional abnormalities favoring a marked pro-inflammatory phenotype and structural imbalances in the intestinal ecosystem, resulting in a decrease in the growth rates of beneficial microorganisms and an increase in the growth rates of certain pathogenic or opportunistic bacteria.
Intestinal microorganisms have recently gained much attention as potential drivers of CD. This idea is supported by the fact that the close interaction between the gut microbiota and the intestinal mucosa continually influences the intestinal immune system, and any imbalance of the normal microbiome state can likely trigger immune dysfunction through pro-inflammatory signals (24).
With the advancement of metagenomic technology, growing evidence suggests that intestinal dysbiosis, or the imbalance in normal gut microbiota, can promote chronic inflammatory conditions and the production of carcinogenic metabolites leading to neoplasia (25).
Although genes contribute to colorectal cancer, the gut microbiota plays a significant role. Scientific evidence suggests that chronic infection and subsequent inflammation contribute to tumor initiation and progression.
Various bacterial species and tumor-promoting virulence mechanisms have been investigated. Significant advances have been made in understanding the composition and functional capabilities of the gut microbiome and its role in cancer (3).
Some bacteria have been consistently increased (Fusobacteria, Alistipes, Porphyromonadaceae, Coriobacteridae, Staphylococcaceae, Akkermansia spp., Methanobacteriales) while others have been consistently decreased in colorectal cancer (Bifidobacterium, Lactobacillus, Ruminococcus, Faecalibacterium spp., Roseburia, Treponema). It has also been shown that bacterial amino acid metabolites are increased, and butyrate is decreased during carcinogenesis (26).
Celiac disease is a digestive disorder affecting approximately 1% of the population, although a much higher percentage carries genes related to genetic susceptibility.
A study demonstrated that the microbiome could be a cofactor in the development of the disease in genetically predisposed people. The study conducted on mice showed that the duodenal microbiome could protect or provoke celiac disease in genetically susceptible mice, depending on the microbial composition and the balance between pathobionts and commensals.
In other words, microorganisms that have the potential to cause diseases if conditions are favorable, and commensals, which are microorganisms that live in symbiosis with the host, benefiting from the relationship without causing harm (27).
Depression is one of the most common mental disorders. However, its causes are still partially unknown, and its diagnosis is complex. A group of Dutch researchers detected variations in the gut microbiome characteristic of individuals with depression, verifying that such variations are independent of the ethnic group.
The study indicates that some of the involved bacteria are producers of metabolites such as glutamate, butyrate, serotonin, and gamma-aminobutyric acid (GABA), which are substances with potent neurological action. These substances would exert their effect through the vagus nerve, which connects the gut and the brain, influencing mood and depression (28).
With technological advancements, knowledge about the gut microbiome has expanded considerably in recent years. The influence of the gut microbiome on health and disease is currently one of the most explored areas of Science.
Deepening our understanding of how disruptions to the early-life gut microbiome, combined with other life stresses, impact health, can provide new insights into the treatment of many complex disorders and diseases (such as fibromyalgia, inflammatory bowel disease, autism).
Future research should focus on the gut microbiome as a biomarker for dietary intake and disease development, the effect of probiotic supplementation on clinical outcomes, and bacterial therapy for intestinal health, immunity, and therapeutic treatments (29).

SYNLAB offers the MyBiome test, a unique diagnostic test of the gut microbiome, as it performs a complete reading of the gut microbial genome through shotgun metagenomics sequencing, allowing for a thorough, objective and actionable study of all microorganisms that make up the intestinal ecosystem, providing analysis:
Thus, MyBiome provides detailed information about the microorganisms inhabiting the gut and their functionality, the impact on health, which key functions for health are being performed by the detected microorganisms, and how to achieve balance through personalized nutritional recommendations.
The MyBiome test is performed using shotgun sequencing, a metagenomic technique that involves the random fragmentation of DNA fragments, followed by the individual sequencing of these fragments.
This approach is highly effective for sequencing whole genomes, especially of organisms whose genome is too large to be sequenced conventionally. Thus, shotgun sequencing enables a comprehensive analysis of the genome of all bacteria and microorganisms present in the intestinal microbiota. Through metagenomic sequencing, comprehensive information about the genome of an organism can be obtained, allowing for the analysis of genes, metabolic pathways, and regulatory regions, as the entire genome is sequenced, which includes millions to billions of base pairs.
In contrast, 16S gene sequencing is employed only to identify and classify bacteria present in a sample, as only a specific region of the 16S rRNA gene, relatively short (about 1,500 bases), is sequenced (30).
The use of metagenomic shotgun sequencing for gut microbiota analysis presents several advantages compared to studies based on 16S gene sequencing or PCR-RT.
Conventional analysis of bacterial populations by amplification of the 16S ribosomal RNA gene (16S rRNA) relies on amplification of hypervariable regions of this gene through primers (base-initiating sequences that complement and specifically bind to regions of interest in DNA), which amplify small regions of the 16S in most bacteria present in a sample.
However, this technique does not allow for the detection of microorganisms that have undergone modifications in the primer binding site (31). Therefore, these bacteria are technically “invisible” and escape analysis (32).
Furthermore, the 16S rRNA gene represents only a small part of a bacterium’s entire genome, making any analysis beyond phylogenetic classification difficult.
Compared to conventional microbiota studies based on 16S rRNA analysis, MyBiome allows for the detection of all genes present in sample microorganisms thanks to the use of metagenomic sequencing technique, avoiding amplification bias and facilitating a comprehensive sampling of all genes present in organisms (33).
The metagenomic approach provides a deeper analysis, providing information not only on all bacteria present down to the species and strain taxonomic level, but also on other microorganisms present in the sample such as fungi, archaea, and protists.
In addition to identification, metagenomic sequencing allows for the characterization and quantification of functional genes, providing information about the functions of microorganisms present in the sample.
The technology used in MyBiome also stands out for its high resolution compared to conventional 16S rRNA gene amplification analysis. While 16S gene sequencing does not allow discrimination between bacterial species with similar or identical 16S regions (34), metagenomic sequencing enables more precise identification of microorganisms and their functional genes.
This limitation implies significant loss of information, as different species within the same genus can have very distinct functions (35). Whole genome shotgun sequencing used in MyBiome is currently the highest resolution technique for identifying microorganisms and detecting their functional genes.
These characteristics, combined with personalized recommendations, make MyBiome a unique analysis in the market, providing detailed insights into intestinal microbiota and its functionality.
MyBiome is especially indicated for:
Performing accurate and up-to-date tests, such as MyBiome, is essential for more accurate diagnoses and better treatment guidance. And SYNLAB is here to help you.
We offer diagnostic solutions with rigorous quality control to the companies, patients, and physicians we serve. We have been in Brazil for over 10 years, operating in 36 countries and three continents, and we are leaders in service provision in Europe.
Contact the SYNLAB team and learn about the available tests.
References
(1) Berg G, et al. Microbiome definition re-visited: old concepts and new challenges. Microbiome. 2020 Jun 30;8(1):103. doi: 10.1186/s40168-020-00875-0.
(2) Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402), 207-214.
(3) Sun J; Kato I. Gut microbiota, inflammation and colorectal câncer. Genes Dis. 2016 Jun;3(2):130-143.
(4) Schoeler Marc; Caesar Robert. Dietary lipids, gut microbiota and lipid metabolismo. Reviews in Endocrine and Metabolic Disorders (2019) 20:461–472.
(5) Atanu Adak; Mojibur R. Khan. An insight into gut microbiota and its functionalities. Cellular and Molecular Life Sciences (2019) 76:473–493.
(6) Sonnenburg JL, Backhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535(7610):56–64.
(7) Sarkar A, Lehto SM, Harty S, Dinan TG, Cryan JF, Burnet PW. (2016) Psychobiotics and the manipulation of bacteria–gut–brain signals. Trends Neurosci 39(11):763–781.
(8) LeBlanc, J.G. et al. (2013) Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr. Opin.Biotechnol. 24, 160–168
(9) Claus, S.P. et al. (2016) The gut microbiota: a major player in the toxicity of environmental pollutants? NPJ Biofilms Microbiomes 2, 16003
(10) Kamada, N. et al. (2013) Role of the gut microbiota inimmunity and inflammatory disease. Nat. Rev.Immunol. 13, 321–335).
(11) Ursell LK, Metcalf JL, Parfrey LW, Knight R. Defning the human microbiome. Nutr Ver. 2012;70(Suppl 1):S38-44.
(12) Hollister, E. B. et al. Structure and function of the healthy pre-adolescent pediatric gut microbiome. Microbiome. 2015;3,36.
(13) Claesson, M. J. et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488,178–184.
(14) Stewart, C. J. et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018;562,583–588.
(15) Arrieta MC, Stiemsma LT, Amenyogbe N, Brown EM, Finlay B. The intestinal microbiome in early life: health and disease. Front. Immunol. 2014;5,427.
(16) Le Chatelier, E. et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500,541–546.
(17) Cryan, J. F., O’Riordan, K. J., Sandhu, K., Peterson, V. & Dinan, T. G. The gut microbiome in neurological disorders. Lancet Neurol. 2020;19,179–194.
(18) Nejman, D. et al. The human tumor microbiome is composed of tumor type specific intracellular bacteria. Science. 2020;368,973–980.
(19) Hepworth, M. R. et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498, 113–117.
(20) Rackaityte E, Lynch SV. The human microbiome in the 21st century. Nat Commun. 2020 Oct 16;11(1):5256. doi: 10.1038/s41467-020-18983-8.
(21) Scharschmidt, T. C. et al. A wave of regulatory T Cells into neonatal skin mediates tolerance to commensal microbes. Immunity. 2015;43,1011–1021.
(22) Qing He, Yuan Gao, Zhuye Jie, Xinlei Yu, Janne Marie Laursen. Two distinct metacommunities characterize the gut microbiota in Crohn’s disease patients. Gigascience. 2017 Jul 1;6(7):1-11.
(23) Katsuyoshi Matsuoka 1, Takanori Kanai. The gut microbiota and inflammatory bowel disease. Semin Immunopathol. 2015 Jan;37(1):47-55.
(24) He Q, et al. Two distinct metacommunities characterize the intestinal microbiota in patients with Crohn’s disease. Gigascience. 2017 Jul 1;6(7):1-11.
(25) Marchesi JR, Dutilh BE, Hall N, et al. Towards the human colorectal cancer microbiome. PLoS One. 2011;6:e20447.
(26) Borges-Canha M, et al. Role of colonic microbiota in colorectal carcinogenesis: a systematic review. Rev Esp Enferm Dig. 2015;107.
(27) Constante M, et al. Biogeographic Variation and functional pathways of the Gut microbiota in Celiac Disease. Gastroenterology 2022 163(5):1351-1363.e15.
(28) Radjabzadeh D, eta l. Gut microbiome-wide association study of depressive symptoms. Nat Commun. 2022. 6;13(1)7128.
(29) Cresci G, Bawden E. The Gut Microbiome: What we do and don’t know. Nutr Clin Pract. 2015 December; 30(6): 734–746.
(30) Ranjan R, Rani A, Metwally A, McGee HS, Perkins DL. Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem Biophys Res Commun. 2016;22;469(4):967-77.
(31) Yang B, Wang Y, Qian PY. Sensitivity and correlation of hypervariable regions in 16S rRNA genes in phylogenetic analysis. BMC Bioinformatics. 2016 Mar 22;17:135.
(32) Větrovský T, Baldrian P. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS One. 2013;8(2):e57923.
(33) Kumar S, Krishnani KK, Bhushan B, Brahmane MP. Metagenomics: Retrospect and Prospects in High Throughput Age. Biotechnol Res Int. 2015;2015:121735.
(34) Fox GE, Wisotzkey JD, Jurtshuk P Jr. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol.1992 Jan;42(1):166-70.
(35) Clarridge JE 3rd. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev. 2004 Oct;17(4):840-62.
Find out about the exam
Serotonin, popularly known as the “happiness hormone,” is a fundamental…
Continue reading
Have you ever felt discomfort after eating certain foods, such…
Continue reading
Dermatomyositis is a rare inflammatory disease that affects the skin…
Continue reading